|
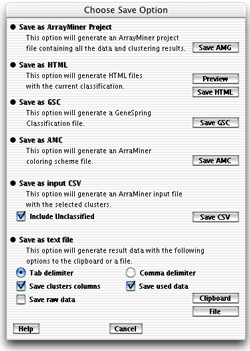
- Clustering
save formats
- Save as ArrayMiner Project (amg
extension). This option will save the solution and the algorithm
status to the disk.
- Save as HTML. This option will
save the current classification as linked html files. Use the "Preview"
button to preview the result in your browser. Or the "Save
HTML" button to specify a target folder for the html
files. The main page will be named "index.html". You are
then free to modify the generated files within your favorite html
software. (Note that the profiles will be sorted in the same way
as specified in the heatmap
view table) More on this topic here
- Save as GeneSpring Classification File
(gsc extension). This
will save the solution as a GeneSpring classification file. To open
it from GeneSpring, launch GeneSpring, choose the appropriate organism,
go to the external program interface and choose the "Load
Classification From File" option.
- Save as ArrayMiner coloring scheme file
(amc extension). This
will save the current solution in a file you can open with the coloring
scheme panel or the classification
compare window
- Save as input CSV. This will generate an input
file for ArrayMiner. This format does not include any classification.
If you uncheck the "Include Unclassified"
checkbox, only the genes assigned to a cluster will be saved. (This
checkbox is relevant only in the gaussian
mode of the clustering)
- Save as text file. This will save
the result in a tabular text file or copy them into clipboard depending
on the chosen button. The data is formatted one gene per line. You
can choose to use tab or commas as cell delimiters. Checking the
Save Clusters Columns checkbox will add a column with the ID or
the name of the assigned cluster. If you check the "Save
used data" checkbox, experiments values seen by the
algorithm will be added to the file (After normalizations).
If you check the "Save raw data" checkbox,
experiments values as readed will be added to the file (Before normalizations)
|