|
|
Saving Clusters in HTML for the Web |
 |
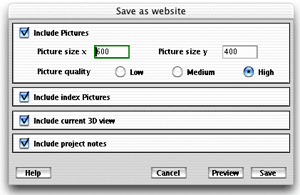
This option of the save
solution dialog box allows you to generate a whole website describing
your current classification. To save the generated files, click on the
"Save" button and specify the output folder.
The output folder will include multiple files :
|
|
Parameters |
|
 |
|
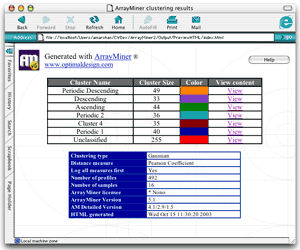
Cluster Selector HTML file (index.html) This html file contains two tables. The first one lets you view the content of the clusters present in the classification and the second one details about the classification parameters. The cluster table
Each line of the table describes a cluster. The view content column gives you access to each cluster content. The information table
This table displays details on the classification :
|
Cluster Content Page You can access these pages by clicking on a "View" link in the cluster selector page which is accessible via the "Back to index" button. The header on the top of the page displays the cluster name and size. The table is filled with all the members of the cluster as shown in the following sample.
|
| Note | The resulting html files can be directly open in most popular spreadsheet software, such as Microsoft Excel®. |