|
|
Using the experiment tree window |
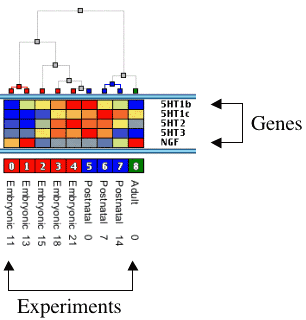 |
Experiment Trees Experiment trees are a convenient way to view how your experiments match together. The tree is built with the following rules :
Each node is colorized by the attribute of the experiment until the node is merged with a node with another attribute. The distance of each node to its two leaves is proportional to their distance. Clicking and maintaining the mouse down on a node display the distance between the two leaves. |
|
Left buttons
Preferences control
|