|
|
The Search Window (Internet Tab) |
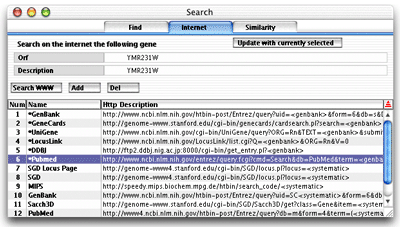 The Search Window (Internet Tab) |
The "Internet"
Tab of the Search window gives you the oppportunity to query gene information
on the Web. To update the orf to be searched, simply click on the "Update
with currently selected" button. The selected gene is marked
with a red target in the 2D and
3D mode. Choose the appropriate
Internet query in the table and click on the "Search WWW"
button. |
| You can add new queries by
hitting the "Add" button. To modify the name
of a query simply double click on its name in the table. To modify the
query description, simply double click on its description in the table.
If your query needs to use the orf of the gene, fill the orf part in the
query with <systematic> as shown in the following
sample. When running the query, ArrayMiner will replace <systematic>
with the orf identifier. If you need to use the GenBank ID of the gene in your query, fill the appropriate part with "<genbank>", as shown in the following example.When running the query, ArrayMiner will replace <genbank> with the GenBank ID of the orf. http://www.ncbi.nlm.nih.gov/cgi-bin/UniGene/query?ORG=Rn&TEXT=<genbank>&submit=Go&ORG=Rn If needed, you can use multiple <genbank> and <systematic> flags in the same query. If you are using GeneSpring and had configured the Internet queries in GeneSpring, you can use the Refresh Annotations option in GeneSpring's external program interface to automatically generate this table (the queries are imported with annotations). This will also fill the GenBank accession numbers if available. You can also directly generate a tsv file (Tab Separated Values) to generate this table as explained in the next section. Creating the Internet query database from a text file : If you are running ArrayMiner as a standalone tool, you need to specify in a tsv file (Tab Separated Values), the engines and queries you want to use, in the following format :
|