|
|
The Classification Compare Window |
| Graphical representation of classifications
The Display Panel can display up to
nine different classifications of the data currently loaded into ArrayMiner.
Each of the classifications is marked on top by the name of the file it
was retrieved from (a red point will designate that this is a file
based classification), or the number of ArrayMiner's iteration if
it comes directly from the current optimization process. The classifications
are automatically ordered from the left to the right according to increasing
number of clusters. |
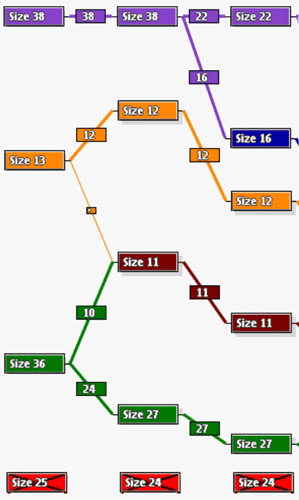 |
Thus in the example in the left figure, the first classification consists of four clusters containing 38, 13, 36 and 25 genes, respectively. The last cluster is a noise cluster. When the same data were classified into five groups, the Classification Compare shows that
Similar correspondences are found when comparing the evolution of classification into six clusters. In order to know exactly what genes are in each cluster and which ones switch clusters among different classifications it is possible to
|