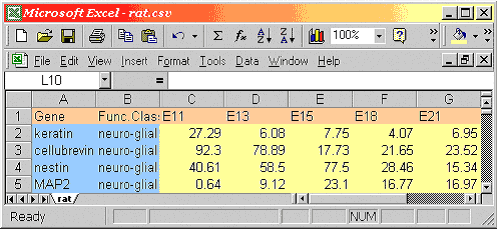
| You can also enter your data in
a text file and save the result as yourfile.csv. For example the following
file text is equivalent to the Excel spreadsheet depicted figure 1:
Gene,Func.Class.1,E11,E13,E15,E18,E21,P0,P7,P14,A
keratin,neuro-glial markers,27.29,6.08,7.75,4.07,6.95,3.42,2.14,0.59,0
cellubrevin,neuro-glial markers,92.3,78.89,17.73,21.65,23.52,18.8,26.07,29.07,9.82
nestin,neuro-glial markers,40.61,58.5,77.5,28.46,15.34,8.32,3.55,3.75,1.59
MAP2,neuro-glial markers,0.64,9.12,23.1,16.77,16.97,11.07,9.58,11.63,6.83
Missing values are represented
by a double comma.
In case of difficulties in formatting
your data, you can use the data importer window
which will automatically convert your data into a format directly readable
by ArrayMiner.
|